Abstract
Introduction: Large Granular Lymphocyte (LGL) leukemia is a rare lymphoproliferative disorder usually manifested in elderly individuals, with mean age of onset at 65. However, LGL leukemia cases in pediatric and young adult (age <30) populations are occasionally observed. Severity of disease, response to routine LGL treatments (methotrexate, cyclophosphamide, cyclosporine), and molecular profiling of this cohort is largely unknown, with only a few case reports describing the occurrence. Here we present the first comprehensive analysis of pediatric and young adult LGL leukemia patients, where we compare demographic and hematologic parameters, frequency of STAT3 mutations, whole exome and transcriptomic profile, and treatment responses to the older adult (age >50) cohort.
Methods: We retrospectively collected clinical data from 23 young adult (age <30), 105 adult (age 30-50), and 89 older adult (age >50) LGL leukemia patients. All participants signed informed consent to provide biospecimens and clinical records to the LGL Leukemia Registry. Whole exome (PBMC and saliva) and RNA sequencing (PBMC) was conducted on 115 patients. Sanger sequencing was utilized to determine STAT3 mutational status in the remaining 102 patients.
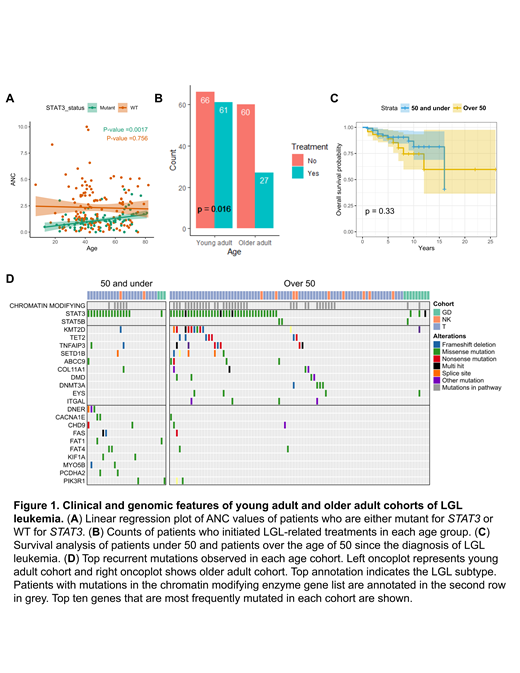
Results: STAT3 is the most frequently mutated gene in LGL leukemia, and the mutation is associated with lower absolute neutrophil counts (ANC). We observed no differences in the frequency of STAT3 somatic mutations in young adult, adult, and older adult LGL leukemia (p=0.143). However, we detected a significant positive linear relationship between age of the patient and ANC values (p=0.002) for STAT3 mutant patients (Figure 1A). STAT3 wild-type (WT) patients did not display this relationship (p=0.756). No age associated trends were detected for hemoglobin values, although we detected decreasing red blood cell (RBC) values with increasing age for both STAT3 mutated and WT patients (p= 0.031, 0.010, respectively). No difference in sex (male vs female) balance was found within the young adult cohort (p=0.190). As rheumatoid arthritis (RA) is observed in ~30% of LGL leukemia patients, we asked if this prevalence also holds in the younger cohort. RA was observed in similar proportion in all young, adult, and older adult groups (p=1).
To compare the severity of disease manifestation between the age groups, we first asked how many patients ever initiated LGL-related treatments in each age group. We found that patients under 50 are more likely to be on LGL related treatments compared to patients over the age of 50 (p=0.016) (Figure 1B). Next, we examined length of survival between age groups. Despite increased initiation of LGL related treatments, no statistically significant difference in overall survival (OS) was observed between patients under 50 and patients over the age of 50 (p=0.33) (Figure 1C). Therefore, we asked if the treatment response in younger patients is better compared to older patients. We observed no statistically significant differences in the routine LGL treatment responses between patients under 50 and patients over the age of 50 (p=0.732).
Interestingly, epigenetic and chromatin modifier mutations are equally common in patients 50 years old and under relative to older adult patients (p= 0.271) (Figure 1D). Transcriptomic analysis of the PBMC data revealed enrichment of TNFα signaling in patients older than 50, while IFNα and IFNγ signaling was enriched in patients 50 years old and younger.
Conclusions: We report the clinical and molecular analysis of LGL leukemia in pediatric/young adult and adult patients compared to the older adult patients that comprise the majority of LGL cases. We show that while STAT3 mutation frequency is similar across all age groups, younger patients with STAT3 mutation show lower ANC values compared to older patients. Importantly, we show that younger patients exhibit a more severe disease, as indicated by increased initiation of LGL treatment. However, overall survival in younger groups compared to older patients is not impacted. Finally, we show that younger patients display similar proportions of epigenetic and chromatin modifying mutations that are a prominent feature of older patients, and feature enrichment of IFNα and IFNγ signaling distinct from older patients.
Feith: Kymera Therapeutics: Membership on an entity's Board of Directors or advisory committees. Loughran: Dren Bio: Membership on an entity's Board of Directors or advisory committees; Keystone Nano: Membership on an entity's Board of Directors or advisory committees; Bioniz Therapeutics: Membership on an entity's Board of Directors or advisory committees; Kymera Therapeutics: Membership on an entity's Board of Directors or advisory committees.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal